Ramp Optimization Examples¶
This notebook outlines an example to optimize the ramp settings for a few different types of observations.
In these types of optimizations, we must consider observations constraints such as saturation levels, SNR requirements, and limits on acquisition time.
Note: The reported acquisition time does not include obsevatory and instrument-level overheads, such as slew times, filter changes, script compilations, etc. It only includes detector readout times (including reset frames).
[1]:
# Import the usual libraries
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
# Enable inline plotting at lower left
%matplotlib inline
[2]:
import pynrc
from pynrc import nrc_utils
from pynrc.nrc_utils import S, jl_poly_fit
from pynrc.pynrc_core import table_filter
pynrc.setup_logging('WARNING', verbose=False)
from astropy.table import Table
# Progress bar
from tqdm.auto import tqdm, trange
Example 1: M-Dwarf companion (imaging vs coronagraphy)¶
We want to observe an M-Dwarf companion (K=18 mag) in the vicinity of a brighter F0V (K=13 mag) in the F430M filter. Assume the M-Dwarf flux is not significantly impacted by the brighter PSF (ie., in the background limited regime). In this scenario, the F0V star will saturate much more quickly compared to the fainter companion, so it limits which ramp settings we can use.
We will test a couple different types of observations (direct imaging vs coronagraphy).
[3]:
# Get stellar spectra and normalize at K-Band
# The stellar_spectrum convenience function creates a Pysynphot spectrum
bp_k = S.ObsBandpass('k')
sp_M2V = pynrc.stellar_spectrum('M2V', 18, 'vegamag', bp_k)#, catname='ck04models')
sp_F0V = pynrc.stellar_spectrum('F0V', 13, 'vegamag', bp_k)#, catname='ck04models')
[4]:
# Initiate a NIRCam observation
nrc = pynrc.NIRCam(filter='F430M', wind_mode='WINDOW', xpix=160, ypix=160)
[5]:
# Set some observing constraints
# Let's assume we want photometry on the primary to calibrate the M-Dwarf for direct imaging
# - Set well_frac_max=0.75
# Want a SNR~100 in the F430M filter
# - Set snr_goal=100
res = nrc.ramp_optimize(sp_M2V, sp_bright=sp_F0V, snr_goal=100, well_frac_max=0.75, verbose=True)
BRIGHT1
BRIGHT2
DEEP2
DEEP8
MEDIUM2
MEDIUM8
RAPID
SHALLOW2
SHALLOW4
Pattern NGRP NINT t_int t_exp t_acq SNR Well eff
---------- ---- ---- --------- --------- --------- -------- -------- --------
DEEP8 7 4 35.67 142.66 143.80 99.3 0.628 8.277
DEEP8 7 5 35.67 178.33 179.75 111.0 0.628 8.277
DEEP2 8 4 39.57 158.27 159.40 98.5 0.696 7.804
DEEP2 8 5 39.57 197.83 199.25 110.2 0.696 7.804
DEEP8 5 7 24.52 171.64 173.63 101.4 0.432 7.697
MEDIUM8 8 8 21.73 173.87 176.14 100.5 0.383 7.575
MEDIUM8 8 9 21.73 195.61 198.16 106.6 0.383 7.575
MEDIUM2 10 7 25.63 179.44 181.43 98.6 0.451 7.322
... ... ... ... ... ... ... ... ...
RAPID 10 555 2.79 1546.45 1703.96 101.5 0.049 2.458
RAPID 10 556 2.79 1549.24 1707.03 101.6 0.049 2.458
RAPID 10 557 2.79 1552.02 1710.10 101.7 0.049 2.458
RAPID 10 558 2.79 1554.81 1713.17 101.7 0.049 2.458
RAPID 10 559 2.79 1557.60 1716.24 101.8 0.049 2.458
RAPID 10 560 2.79 1560.38 1719.31 101.9 0.049 2.458
RAPID 10 561 2.79 1563.17 1722.38 102.0 0.049 2.458
RAPID 8 1046 2.23 2331.66 2628.51 102.0 0.039 1.990
Length = 55 rows
[6]:
# Print the Top 2 settings for each readout pattern
res2 = table_filter(res, 2)
print(res2)
Pattern NGRP NINT t_int t_exp t_acq SNR Well eff
---------- ---- ---- --------- --------- --------- -------- -------- --------
RAPID 10 539 2.79 1501.87 1654.84 100.0 0.049 2.458
RAPID 10 540 2.79 1504.66 1657.91 100.1 0.049 2.458
BRIGHT1 10 145 5.29 767.65 808.80 99.8 0.093 3.509
BRIGHT1 10 146 5.29 772.95 814.38 100.1 0.093 3.509
BRIGHT2 10 93 5.57 518.27 544.66 99.7 0.098 4.270
BRIGHT2 10 94 5.57 523.84 550.52 100.2 0.098 4.270
SHALLOW2 10 20 13.10 261.92 267.60 99.6 0.230 6.089
SHALLOW2 10 21 13.10 275.02 280.98 102.1 0.230 6.089
SHALLOW4 10 16 13.65 218.45 222.99 99.7 0.240 6.675
SHALLOW4 10 17 13.65 232.11 236.93 102.8 0.240 6.675
MEDIUM2 10 7 25.63 179.44 181.43 98.6 0.451 7.322
MEDIUM2 10 8 25.63 205.08 207.35 105.4 0.451 7.322
MEDIUM8 8 8 21.73 173.87 176.14 100.5 0.383 7.575
MEDIUM8 8 9 21.73 195.61 198.16 106.6 0.383 7.575
DEEP2 8 4 39.57 158.27 159.40 98.5 0.696 7.804
DEEP2 8 5 39.57 197.83 199.25 110.2 0.696 7.804
DEEP8 7 4 35.67 142.66 143.80 99.3 0.628 8.277
DEEP8 7 5 35.67 178.33 179.75 111.0 0.628 8.277
[7]:
# Do the same thing, but for coronagraphic mask instead
nrc = pynrc.NIRCam(filter='F430M', image_mask='MASK430R', pupil_mask='CIRCLYOT',
wind_mode='WINDOW', xpix=320, ypix=320)
# We assume that longer ramps will give us the best SNR for time
patterns = ['MEDIUM8', 'DEEP8']
res = nrc.ramp_optimize(sp_M2V, sp_bright=sp_F0V, snr_goal=100,
patterns=patterns, even_nints=True)
# Take the Top 2 settings for each readout pattern
res2 = table_filter(res, 2)
print(res2)
Pattern NGRP NINT t_int t_exp t_acq SNR Well eff
---------- ---- ---- --------- --------- --------- -------- -------- --------
MEDIUM8 10 94 104.77 9848.00 9950.36 99.6 0.001 0.998
MEDIUM8 10 96 104.77 10057.53 10162.07 100.7 0.001 0.998
DEEP8 20 14 414.79 5807.03 5822.27 104.6 0.003 1.370
DEEP8 19 14 393.41 5507.69 5522.94 101.2 0.003 1.362
RESULTS
Based on these two comparisons, it looks like direct imaging is much more efficient in getting to the requisite SNR. In addition, direct imaging gives us a photometric comparison source that is inaccessible when occulting the primary with the coronagraph masks. Of course, this assumes the companion exists in the background limit as opposed to the contrast limit.
Example 2: Exoplanet Coronagraphy¶
We want to observe GJ 504 for an hour in the F444W filter using the MASK430R coronagraph. - What is the optimal ramp settings to maximize the SNR of GJ 504b? - What is the final background sensitivity limit?
[8]:
# Get stellar spectra and normalize at K-Band
# The stellar_spectrum convenience function creates a Pysynphot spectrum
bp_k = pynrc.bp_2mass('ks')
sp_G0V = pynrc.stellar_spectrum('G0V', 4, 'vegamag', bp_k)
# Choose a representative planet spectrum
planet = pynrc.planets_sb12(atmo='hy3s', mass=8, age=200, entropy=8, distance=17.5)
sp_pl = planet.export_pysynphot()
# Renormalize to F360M = 18.8
bp_l = pynrc.read_filter('F360M') #
sp_pl = sp_pl.renorm(18.8, 'vegamag', bp_l)
[9]:
# Initiate a NIRCam observation
nrc = pynrc.NIRCam(filter='F444W', pupil_mask='CIRCLYOT', image_mask='MASK430R',
wind_mode='WINDOW', xpix=320, ypix=320)
[10]:
# Set even_nints=True assume 2 roll angles
res = nrc.ramp_optimize(sp_pl, sp_bright=sp_G0V, tacq_max=3600, tacq_frac=0.05,
even_nints=True, verbose=True)
BRIGHT1
BRIGHT2
DEEP2
DEEP8
MEDIUM2
MEDIUM8
RAPID
SHALLOW2
SHALLOW4
Pattern NGRP NINT t_int t_exp t_acq SNR Well eff
---------- ---- ---- --------- --------- --------- -------- -------- --------
DEEP2 17 10 344.23 3442.31 3453.20 672.8 0.777 11.448
DEEP2 16 10 322.85 3228.50 3239.39 651.5 0.728 11.446
DEEP2 16 12 322.85 3874.20 3887.27 713.7 0.728 11.446
DEEP2 15 12 301.47 3617.63 3630.70 689.4 0.680 11.440
DEEP2 14 12 280.09 3361.06 3374.13 664.0 0.632 11.431
DEEP8 17 10 350.65 3506.45 3517.34 676.2 0.791 11.402
DEEP8 16 10 329.26 3292.64 3303.53 655.3 0.743 11.401
DEEP8 15 12 307.88 3694.60 3707.67 694.1 0.695 11.398
... ... ... ... ... ... ... ... ...
BRIGHT2 10 162 21.38 3463.69 3640.10 518.1 0.048 8.587
BRIGHT1 10 166 20.31 3371.75 3552.52 463.1 0.046 7.768
BRIGHT1 10 168 20.31 3412.38 3595.32 465.8 0.046 7.768
BRIGHT1 10 170 20.31 3453.00 3638.12 468.6 0.046 7.768
RAPID 10 304 10.69 3249.88 3580.93 358.9 0.024 5.997
RAPID 10 306 10.69 3271.26 3604.48 360.1 0.024 5.997
RAPID 10 308 10.69 3292.64 3628.04 361.2 0.024 5.997
RAPID 9 334 9.62 3213.53 3577.25 331.8 0.022 5.548
Length = 36 rows
[11]:
# Take the Top 2 settings for each readout pattern
res2 = table_filter(res, 2)
print(res2)
Pattern NGRP NINT t_int t_exp t_acq SNR Well eff
---------- ---- ---- --------- --------- --------- -------- -------- --------
RAPID 10 304 10.69 3249.88 3580.93 358.9 0.024 5.997
RAPID 10 306 10.69 3271.26 3604.48 360.1 0.024 5.997
BRIGHT1 10 166 20.31 3371.75 3552.52 463.1 0.046 7.768
BRIGHT1 10 168 20.31 3412.38 3595.32 465.8 0.046 7.768
BRIGHT2 10 158 21.38 3378.17 3550.22 511.7 0.048 8.587
BRIGHT2 10 160 21.38 3420.93 3595.16 514.9 0.048 8.587
SHALLOW2 10 68 50.24 3416.65 3490.70 606.1 0.113 10.258
SHALLOW2 10 70 50.24 3517.14 3593.37 615.0 0.113 10.258
SHALLOW4 10 66 52.38 3457.28 3529.15 619.9 0.118 10.434
SHALLOW4 10 68 52.38 3562.04 3636.09 629.2 0.118 10.434
MEDIUM2 10 34 98.35 3343.96 3380.98 638.0 0.222 10.971
MEDIUM2 10 36 98.35 3540.66 3579.86 656.5 0.222 10.971
MEDIUM8 10 32 104.77 3352.51 3387.36 638.0 0.236 10.962
MEDIUM8 10 34 104.77 3562.04 3599.07 657.7 0.236 10.962
DEEP2 17 10 344.23 3442.31 3453.20 672.8 0.777 11.448
DEEP2 16 10 322.85 3228.50 3239.39 651.5 0.728 11.446
DEEP8 17 10 350.65 3506.45 3517.34 676.2 0.791 11.402
DEEP8 16 10 329.26 3292.64 3303.53 655.3 0.743 11.401
[12]:
# The SHALLOWs, DEEPs, and MEDIUMs are very similar for SNR and efficiency.
# Let's go with SHALLOW2 for more GROUPS & INTS
# MEDIUM8 would be fine as well.
nrc.update_detectors(read_mode='SHALLOW2', ngroup=10, nint=70)
keys = list(nrc.multiaccum_times.keys())
keys.sort()
for k in keys:
print("{:<10}: {: 12.5f}".format(k, nrc.multiaccum_times[k]))
t_acq : 3593.36880
t_exp : 3517.14160
t_frame : 1.06904
t_group : 5.34520
t_int : 50.24488
t_int_tot1: 51.33384
t_int_tot2: 51.33384
[13]:
# Background sensitivity (5 sigma)
sens_dict = nrc.sensitivity(nsig=5, units='vegamag', verbose=True)
Point Source Sensitivity (5-sigma): 21.43 vegamag
Surface Brightness Sensitivity (5-sigma): 22.74 vegamag/arcsec^2
Example 3: Single-Object Grism Spectroscopy¶
Similar to the above, but instead we want to obtain a slitless grism spectrum of a K=12 mag M0V dwarf. Each grism resolution element should have SNR~100.
[14]:
# M0V star normalized to K=12 mags
bp_k = S.ObsBandpass('k')
sp_M0V = pynrc.stellar_spectrum('M0V', 12, 'vegamag', bp_k)
[15]:
nrc = pynrc.NIRCam(filter='F444W', pupil_mask='GRISMR', wind_mode='STRIPE', ypix=128)
[16]:
# Set a minimum of 10 integrations to be robust against cosmic rays
# Also set a minimum of 10 groups for good ramp sampling
res = nrc.ramp_optimize(sp_M0V, snr_goal=100, nint_min=10, ng_min=10, verbose=True)
BRIGHT1
BRIGHT2
DEEP2
DEEP8
MEDIUM2
MEDIUM8
RAPID
SHALLOW2
SHALLOW4
Pattern NGRP NINT t_int t_exp t_acq SNR Well eff
---------- ---- ---- --------- --------- --------- -------- -------- --------
DEEP2 10 10 123.03 1230.27 1237.08 332.4 0.071 9.449
DEEP8 10 10 127.08 1270.82 1277.64 336.0 0.074 9.399
MEDIUM2 10 10 62.19 621.89 628.71 231.2 0.036 9.219
MEDIUM8 10 10 66.25 662.45 669.27 236.1 0.038 9.127
SHALLOW4 10 10 33.12 331.23 338.04 161.8 0.019 8.802
SHALLOW2 10 10 31.77 317.71 324.52 157.6 0.018 8.746
BRIGHT2 10 12 13.52 162.23 170.41 98.8 0.008 7.567
BRIGHT2 10 13 13.52 175.75 184.61 102.8 0.008 7.567
BRIGHT1 10 15 12.84 192.65 202.87 100.0 0.007 7.021
BRIGHT1 10 16 12.84 205.49 216.40 103.3 0.007 7.021
RAPID 10 42 6.76 283.91 312.52 99.0 0.004 5.599
RAPID 10 43 6.76 290.67 319.96 100.2 0.004 5.599
RAPID 10 44 6.76 297.43 327.41 101.3 0.004 5.599
RAPID 10 45 6.76 304.19 334.85 102.5 0.004 5.599
[17]:
# Print the Top 2 settings for each readout pattern
res2 = table_filter(res, 2)
print(res2)
Pattern NGRP NINT t_int t_exp t_acq SNR Well eff
---------- ---- ---- --------- --------- --------- -------- -------- --------
RAPID 10 42 6.76 283.91 312.52 99.0 0.004 5.599
RAPID 10 43 6.76 290.67 319.96 100.2 0.004 5.599
BRIGHT1 10 15 12.84 192.65 202.87 100.0 0.007 7.021
BRIGHT1 10 16 12.84 205.49 216.40 103.3 0.007 7.021
BRIGHT2 10 12 13.52 162.23 170.41 98.8 0.008 7.567
BRIGHT2 10 13 13.52 175.75 184.61 102.8 0.008 7.567
SHALLOW2 10 10 31.77 317.71 324.52 157.6 0.018 8.746
SHALLOW4 10 10 33.12 331.23 338.04 161.8 0.019 8.802
MEDIUM2 10 10 62.19 621.89 628.71 231.2 0.036 9.219
MEDIUM8 10 10 66.25 662.45 669.27 236.1 0.038 9.127
DEEP2 10 10 123.03 1230.27 1237.08 332.4 0.071 9.449
DEEP8 10 10 127.08 1270.82 1277.64 336.0 0.074 9.399
[18]:
# Let's say we choose SHALLOW4, NGRP=10, NINT=10
# Update detector readout
nrc.update_detectors(read_mode='SHALLOW4', ngroup=10, nint=10)
keys = list(nrc.multiaccum_times.keys())
keys.sort()
for k in keys:
print("{:<10}: {: 12.5f}".format(k, nrc.multiaccum_times[k]))
t_acq : 338.04264
t_exp : 331.22530
t_frame : 0.67597
t_group : 3.37985
t_int : 33.12253
t_int_tot1: 33.80374
t_int_tot2: 33.80374
[19]:
# Print final wavelength-dependent SNR
# For spectroscopy, the snr_goal is the median over the bandpass
snr_dict = nrc.sensitivity(sp=sp_M0V, forwardSNR=True, units='mJy', verbose=True)
F444W SNR for M0V source
Wave SNR Flux (mJy)
--------- --------- ----------
3.80 9.52 4.54
3.90 230.66 4.34
4.00 246.18 4.15
4.10 235.68 3.97
4.20 221.71 3.73
4.30 197.92 3.26
4.40 184.55 3.19
4.50 169.03 2.85
4.60 154.66 2.79
4.70 139.31 2.63
4.80 131.21 2.70
4.90 115.05 2.60
5.00 57.52 2.38
5.10 1.30 2.46
Mock observed spectrum
Create a series of ramp integrations based on the current NIRCam settings. The gen_exposures() function creates a series of mock observations in raw DMS format by default. By default, it’s point source objects centered in the observing window.
[20]:
# Ideal spectrum and wavelength solution
wspec, imspec = nrc.calc_psf_from_coeff(sp=sp_M0V, return_hdul=False, return_oversample=False)
# Resize to detector window
nx = nrc.det_info['xpix']
ny = nrc.det_info['ypix']
# Shrink/expand nx (fill value of 0)
# Then shrink to a size excluding wspec=0
# This assumes simulated spectrum is centered
imspec = nrc_utils.pad_or_cut_to_size(imspec, (ny,nx))
wspec = nrc_utils.pad_or_cut_to_size(wspec, nx)
[21]:
# Add simple zodiacal background
im_slope = imspec + nrc.bg_zodi()
[22]:
# Create a series of ramp integrations based on the current NIRCam settings
# Output is a single HDUList with 10 INTs
# Ignore detector non-linearity to return output in e-/sec
kwargs = {
'apply_nonlinearity' : False,
'apply_flats' : False,
}
res = nrc.simulate_level1b('M0V Target', 0, 0, '2023-01-01', '12:00:00',
im_slope=im_slope, return_hdul=True, **kwargs)
[ pynrc:WARNING] Source RA, Dec = (0.0, 0.0) deg is not visible on 2023-01-01!
[23]:
res.info()
Filename: (No file associated with this HDUList)
No. Name Ver Type Cards Dimensions Format
0 PRIMARY 1 PrimaryHDU 117 ()
1 SCI 1 ImageHDU 44 (2048, 128, 10, 10) uint16
2 ZEROFRAME 1 ImageHDU 12 (2048, 128, 10) uint16
3 GROUP 1 BinTableHDU 36 100R x 13C ['I', 'I', 'I', 'J', 'I', '26A', 'I', 'I', 'I', 'I', '36A', 'D', 'D']
4 INT_TIMES 1 BinTableHDU 24 10R x 7C ['J', 'D', 'D', 'D', 'D', 'D', 'D']
[24]:
tvals = nrc.Detector.times_group_avg
header = res['PRIMARY'].header
data_all = res['SCI'].data
slope_list = []
for data in tqdm(data_all):
ref = pynrc.ref_pixels.NRC_refs(data, header, DMS=True, do_all=False)
ref.calc_avg_amps()
ref.correct_amp_refs()
# Linear fit to determine slope image
cf = jl_poly_fit(tvals, ref.data, deg=1)
slope_list.append(cf[1])
# Create a master averaged slope image
slopes_all = np.array(slope_list)
slope_sim = slopes_all.mean(axis=0) * nrc.Detector.gain
[25]:
fig, ax = plt.subplots(1,1, figsize=(12,3))
ax.imshow(slope_sim, vmin=0, vmax=10)
fig.tight_layout()

[26]:
ind = wspec>0
# Estimate background emission and subtract from slope_sim
bg = np.median(slope_sim[:,~ind])
slope_sim -= bg
[27]:
ind = wspec>0
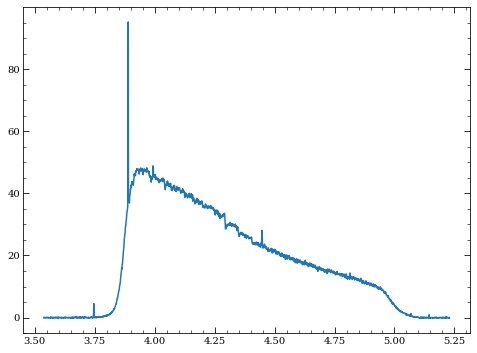
plt.plot(wspec[ind], slope_sim[63,ind])
[27]:
[<matplotlib.lines.Line2D at 0x7fc309945150>]
[28]:
# Extract 2 spectral x 5 spatial pixels
# First, cut out the central 5 pixels
wspec_sub = wspec[ind]
sh_new = (5, len(wspec_sub))
slope_sub = nrc_utils.pad_or_cut_to_size(slope_sim, sh_new)
slope_sub_ideal = nrc_utils.pad_or_cut_to_size(imspec, sh_new)
# Sum along the spatial axis
spec = slope_sub.sum(axis=0)
spec_ideal = slope_sub_ideal.sum(axis=0)
spec_ideal_rebin = nrc_utils.frebin(spec_ideal, scale=0.5, total=False)
# Build a quick RSRF from extracted ideal spectral slope
sp_M0V.convert('mjy')
rsrf = spec_ideal / sp_M0V.sample(wspec_sub*1e4)
# Rebin along spectral direction
wspec_rebin = nrc_utils.frebin(wspec_sub, scale=0.5, total=False)
spec_rebin_cal = nrc_utils.frebin(spec/rsrf, scale=0.5, total=False)
[29]:
# Expected noise per extraction element
snr_interp = np.interp(wspec_rebin, snr_dict['wave'], snr_dict['snr'])
_spec_rebin = spec_ideal_rebin / snr_interp
_spec_rebin_cal = _spec_rebin / nrc_utils.frebin(rsrf, scale=0.5, total=False)
[30]:
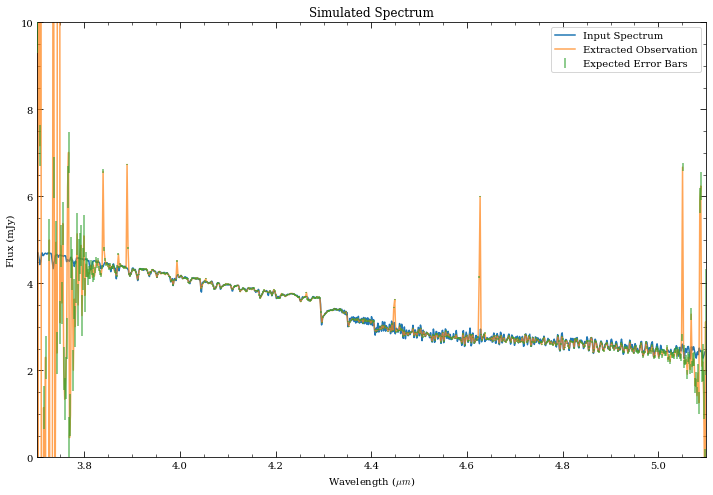
fig, ax = plt.subplots(1,1, figsize=(12,8))
ax.plot(sp_M0V.wave/1e4, sp_M0V.flux, label='Input Spectrum')
ax.plot(wspec_rebin, spec_rebin_cal, alpha=0.7, label='Extracted Observation')
ax.errorbar(wspec_rebin, spec_rebin_cal, yerr=_spec_rebin_cal, zorder=3,
fmt='none', label='Expected Error Bars', alpha=0.7, color='C2')
ax.set_ylim([0,10])
ax.set_xlim([3.7,5.1])
ax.set_xlabel('Wavelength ($\mu m$)')
ax.set_ylabel('Flux (mJy)')
ax.set_title('Simulated Spectrum')
ax.legend(loc='upper right');
Example 4: Exoplanet Transit Spectroscopy¶
Let’s say we want to observe an exoplanet transit using NIRCam grisms in the F322W2 filter.
We assume a 2.1-hour transit duration for a K6V star (K=8.4 mag).
[31]:
nrc = pynrc.NIRCam('F322W2', pupil_mask='GRISM0', wind_mode='STRIPE', ypix=64)
[32]:
# K6V star at K=8.4 mags
bp_k = S.ObsBandpass('k')
sp_K6V = pynrc.stellar_spectrum('K6V', 8.4, 'vegamag', bp_k)
[33]:
# Constraints
well = 0.5 # Keep well below 50% full
tacq = 2.1*3600. # 2.1 hour transit duration
ng_max = 30 # Transit spectroscopy allows for up to 30 groups per integrations
nint_max = int(1e6) # Effectively no limit on number of integrations
# Let's bin the spectrum to R~100
# dw_bin is a passable parameter for specifiying spectral bin sizes
R = 100
dw_bin = (nrc.bandpass.avgwave() / 10000) / R
[34]:
res = nrc.ramp_optimize(sp_K6V, tacq_max=tacq, nint_max=nint_max,
ng_min=10, ng_max=ng_max, well_frac_max=well,
dw_bin=dw_bin, verbose=True)
BRIGHT1
BRIGHT2
DEEP2
DEEP8
MEDIUM2
MEDIUM8
RAPID
SHALLOW2
SHALLOW4
Pattern NGRP NINT t_int t_exp t_acq SNR Well eff
---------- ---- ---- --------- --------- --------- -------- -------- --------
BRIGHT1 24 460 16.01 7363.99 7523.08 30955.5 0.494 356.894
BRIGHT1 24 461 16.01 7380.00 7539.44 30989.1 0.494 356.894
BRIGHT1 24 462 16.01 7396.01 7555.79 31022.7 0.494 356.894
BRIGHT1 24 463 16.01 7412.01 7572.15 31056.3 0.494 356.894
BRIGHT1 24 464 16.01 7428.02 7588.50 31089.8 0.494 356.894
BRIGHT2 23 470 15.67 7363.99 7526.54 30623.8 0.484 352.989
BRIGHT2 23 471 15.67 7379.66 7542.56 30656.4 0.484 352.989
BRIGHT2 23 472 15.67 7395.32 7558.57 30688.9 0.484 352.989
... ... ... ... ... ... ... ... ...
SHALLOW2 10 462 16.01 7396.01 7555.79 30678.0 0.494 352.929
SHALLOW2 10 463 16.01 7412.01 7572.15 30711.2 0.494 352.929
SHALLOW2 10 464 16.01 7428.02 7588.50 30744.4 0.494 352.929
RAPID 30 713 10.22 7285.65 7532.24 30585.0 0.315 352.408
RAPID 30 714 10.22 7295.87 7542.81 30606.5 0.315 352.408
RAPID 30 715 10.22 7306.08 7553.37 30627.9 0.315 352.408
RAPID 30 716 10.22 7316.30 7563.94 30649.3 0.315 352.408
RAPID 30 717 10.22 7326.52 7574.50 30670.7 0.315 352.408
Length = 20 rows
[35]:
# Print the Top 2 settings for each readout pattern
res2 = table_filter(res, 2)
print(res2)
Pattern NGRP NINT t_int t_exp t_acq SNR Well eff
---------- ---- ---- --------- --------- --------- -------- -------- --------
RAPID 30 713 10.22 7285.65 7532.24 30585.0 0.315 352.408
RAPID 30 714 10.22 7295.87 7542.81 30606.5 0.315 352.408
BRIGHT1 24 460 16.01 7363.99 7523.08 30955.5 0.494 356.894
BRIGHT1 24 461 16.01 7380.00 7539.44 30989.1 0.494 356.894
BRIGHT2 23 470 15.67 7363.99 7526.54 30623.8 0.484 352.989
BRIGHT2 23 471 15.67 7379.66 7542.56 30656.4 0.484 352.989
SHALLOW2 10 460 16.01 7363.99 7523.08 30611.6 0.494 352.929
SHALLOW2 10 461 16.01 7380.00 7539.44 30644.8 0.494 352.929
[36]:
# Even though BRIGHT1 has a slight efficiency preference over RAPID
# and BRIGHT2, we decide to choose RAPID, because we are convinced
# that saving all data (and no coadding) is a better option.
# If APT informs you that the data rates or total data shorage is
# an issue, you can select one of the other options.
# Update to RAPID, ngroup=30, nint=700 and plot PPM
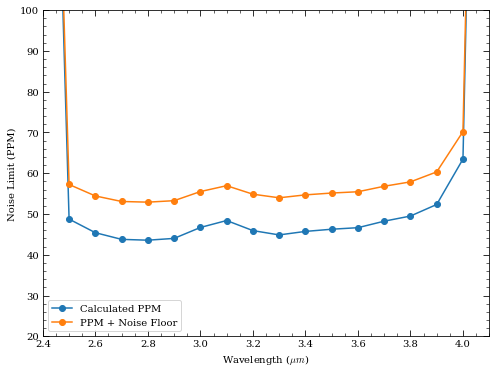
nrc.update_detectors(read_mode='RAPID', ngroup=30, nint=700)
snr_dict = nrc.sensitivity(sp=sp_K6V, dw_bin=dw_bin, forwardSNR=True, units='Jy')
wave = np.array(snr_dict['wave'])
snr = np.array(snr_dict['snr'])
# Let assume bg subtraction of something with similar noise
snr /= np.sqrt(2.)
ppm = 1e6 / snr
# NOTE: We have up until now neglected to include a "noise floor"
# which represents the expected minimum achievable ppm from
# unknown systematics. To first order, this can be added in
# quadrature to the calculated PPM.
noise_floor = 30 # in ppm
ppm_floor = np.sqrt(ppm**2 + noise_floor**2)
[37]:
plt.plot(wave, ppm, marker='o', label='Calculated PPM')
plt.plot(wave, ppm_floor, marker='o', label='PPM + Noise Floor')
plt.xlabel('Wavelength ($\mu m$)')
plt.ylabel('Noise Limit (PPM)')
plt.xlim([2.4,4.1])
plt.ylim([20,100])
plt.legend()
[37]:
<matplotlib.legend.Legend at 0x7fc2f96f0590>
Example 5: Extended Souce¶
Expect some faint galaxies of 25 ABMag/arcsec^2 in our field. What is the best we can do with 10,000 seconds of acquisition time?
[38]:
# Detection bandpass is F200W
nrc = pynrc.NIRCam(filter='F200W')
# Flat spectrum (in photlam) with ABMag = 25 in the NIRCam bandpass
sp = pynrc.stellar_spectrum('flat', 25, 'abmag', nrc.bandpass)
[39]:
res = nrc.ramp_optimize(sp, is_extended=True, tacq_max=10000, tacq_frac=0.05, verbose=True)
BRIGHT1
BRIGHT2
DEEP2
DEEP8
MEDIUM2
MEDIUM8
RAPID
SHALLOW2
SHALLOW4
Pattern NGRP NINT t_int t_exp t_acq SNR Well eff
---------- ---- ---- --------- --------- --------- -------- -------- --------
MEDIUM8 10 9 1052.20 9469.83 9555.73 10.5 0.020 0.107
MEDIUM8 10 10 1052.20 10522.03 10618.67 11.1 0.020 0.107
DEEP8 8 5 1589.04 7945.21 7988.16 9.5 0.031 0.106
DEEP8 7 6 1374.31 8245.84 8299.53 9.7 0.026 0.106
MEDIUM8 8 11 837.47 9212.15 9319.52 10.2 0.016 0.106
DEEP8 6 8 1159.57 9276.57 9351.73 10.3 0.022 0.106
MEDIUM8 9 10 944.84 9448.36 9544.99 10.4 0.018 0.106
DEEP8 8 6 1589.04 9534.25 9587.94 10.4 0.031 0.106
... ... ... ... ... ... ... ... ...
BRIGHT1 10 49 204.00 9995.93 10511.30 6.6 0.004 0.064
BRIGHT1 10 50 204.00 10199.93 10726.04 6.7 0.004 0.064
BRIGHT1 10 51 204.00 10403.93 10940.77 6.8 0.004 0.064
RAPID 10 91 107.37 9770.46 10736.78 4.9 0.002 0.047
RAPID 10 92 107.37 9877.83 10854.88 5.0 0.002 0.047
RAPID 10 93 107.37 9985.20 10972.98 5.0 0.002 0.047
RAPID 10 94 107.37 10092.56 11091.09 5.0 0.002 0.047
RAPID 10 95 107.37 10199.93 11209.19 5.0 0.002 0.047
Length = 66 rows
[40]:
# Print the Top 2 settings for each readout pattern
res2 = table_filter(res, 2)
print(res2)
Pattern NGRP NINT t_int t_exp t_acq SNR Well eff
---------- ---- ---- --------- --------- --------- -------- -------- --------
RAPID 10 91 107.37 9770.46 10736.78 4.9 0.002 0.047
RAPID 10 92 107.37 9877.83 10854.88 5.0 0.002 0.047
BRIGHT1 10 47 204.00 9587.94 10081.83 6.5 0.004 0.064
BRIGHT1 10 48 204.00 9791.93 10296.57 6.6 0.004 0.064
BRIGHT2 10 45 214.74 9663.09 10135.52 7.8 0.004 0.077
BRIGHT2 10 46 214.74 9877.83 10360.99 7.9 0.004 0.077
SHALLOW2 10 18 504.63 9083.31 9265.84 9.3 0.010 0.096
SHALLOW2 10 19 504.63 9587.94 9781.20 9.6 0.010 0.096
SHALLOW4 10 18 526.10 9469.83 9652.36 10.1 0.010 0.102
SHALLOW4 10 19 526.10 9995.93 10189.20 10.3 0.010 0.102
MEDIUM2 10 9 987.78 8890.05 8975.94 9.8 0.019 0.103
MEDIUM2 10 10 987.78 9877.83 9974.46 10.4 0.019 0.103
MEDIUM8 10 9 1052.20 9469.83 9555.73 10.5 0.020 0.107
MEDIUM8 10 10 1052.20 10522.03 10618.67 11.1 0.020 0.107
DEEP2 9 5 1739.36 8696.78 8739.74 9.6 0.033 0.103
DEEP2 8 6 1524.62 9147.73 9201.42 9.9 0.029 0.103
DEEP8 8 5 1589.04 7945.21 7988.16 9.5 0.031 0.106
DEEP8 7 6 1374.31 8245.84 8299.53 9.7 0.026 0.106
[41]:
# MEDIUM8 10 10 looks like a good option
nrc.update_detectors(read_mode='MEDIUM8', ngroup=10, nint=10, verbose=True)
New Ramp Settings
read_mode : MEDIUM8
nf : 8
nd2 : 2
ngroup : 10
nint : 10
New Detector Settings
wind_mode : FULL
xpix : 2048
ypix : 2048
x0 : 0
y0 : 0
New Ramp Times
t_group : 107.368
t_frame : 10.737
t_int : 1052.203
t_int_tot1 : 1052.203
t_int_tot2 : 1062.940
t_exp : 10522.035
t_acq : 10618.671
[42]:
# Calculate flux/mag for various nsigma detection limits
tbl = Table(names=('Sigma', 'Point (nJy)', 'Extended (nJy/asec^2)',
'Point (AB Mag)', 'Extended (AB Mag/asec^2)'))
tbl['Sigma'].format = '.0f'
for k in tbl.keys()[1:]:
tbl[k].format = '.2f'
for sig in [1,3,5,10]:
snr_dict1 = nrc.sensitivity(nsig=sig, units='nJy', verbose=False)
snr_dict2 = nrc.sensitivity(nsig=sig, units='abmag', verbose=False)
tbl.add_row([sig, snr_dict1[0]['sensitivity'], snr_dict1[1]['sensitivity'],
snr_dict2[0]['sensitivity'], snr_dict2[1]['sensitivity']])
[43]:
tbl
[43]:
| Sigma | Point (nJy) | Extended (nJy/asec^2) | Point (AB Mag) | Extended (AB Mag/asec^2) |
|---|---|---|---|---|
| float64 | float64 | float64 | float64 | float64 |
| 1 | 1.10 | 32.05 | 31.30 | 27.64 |
| 3 | 3.30 | 96.57 | 30.10 | 26.44 |
| 5 | 5.52 | 161.67 | 29.55 | 25.88 |
| 10 | 11.14 | 326.99 | 28.78 | 25.11 |
[ ]: